Peptides
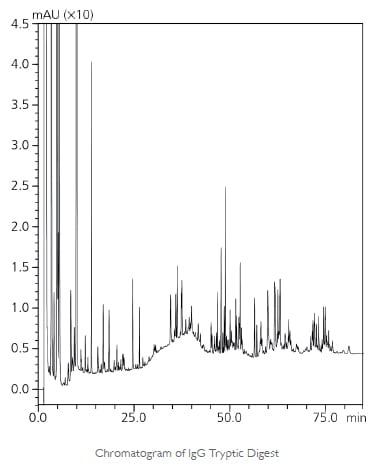
During process development and before entering clinical trials, the primary structure of a recombinant protein must be confirmed, and all potential PTMs must be characterized to assess the associated risk. Peptide mapping is the fundamental technique for this purpose, whereby the recombinant protein is enzymatically digested into peptide fragments that are chromatographically separated to give a fingerprint of the primary structure. When coupled to mass spectrometry, peptide mapping can also give precise identification of various PTMs, including oxidation, deamidation, disulfide bond scrambling, C-terminal lysine truncation and N-terminal pyroglutamination. Shimadzu offers a comprehensive portfolio of solutions for the highly accurate confirmation of protein sequence, identification of modifications, and routine protein fingerprint monitoring for QA/ QC, using Part 11 compliant liquid chromatography and LC/MS systems.




